By: Andrew Oliver | 04/15/2023 SCINet Fellow, Western Human Nutrition Research Center, Davis, CA

Figure 1. Taxonomic relationships of foods consumed in a USDA cohort
Researchers at the USDA-ARS Western Human Nutrition Research Center (WHNRC) are broadly interested in how diet impacts human health. Dr. Danielle Lemay, a Research Molecular Biologist in the Immunity and Disease Prevention Research Unit, together with SCINet Postdoctoral Fellow Dr. Andrew Oliver contribute to this research theme by leveraging machine learning to uncover the molecular underpinnings of diet’s association with health. Of particular interest is the role of the human gut microbiota – a vast and diet-responsive community of microorganisms that live inside our digestive tract and contribute to health through production of beneficial metabolites and commensal interactions with our immune system.
One challenging aspect of investigating the diet-microbiome-health axis is how highly dimensional the data are. For example, a person may consume dozens of different foods in a week, which move through a digestive tract containing thousands of different microbial species, which collectively employ millions of genes to make or modify hundreds of metabolites found in fecal samples. These combinations are highly personalized; indeed, no two individuals share the same gut microbes.
To reduce the dimensionality of the problem, Dr. Oliver developed an algorithmic approach to leverage a property of microbiome data: microbial taxonomy. A common question in microbiome analyses is, “Which microbial species or genera co-vary with my treatment or physiological trait of interest?” This question can be problematic for several reasons; specifically, it 1) assumes the microbiome variability associated with a trait is conserved at the taxonomic level analyzed (e.g., the species or genus) and 2) forces an analysis that can handle hundreds to thousands of (likely) zero-inflated features. Dr. Oliver’s program, called taxaHFE (for taxonomic hierarchical feature envineering), examines every taxonomic level of each feature and determines if it adds discriminatory value, collapsing features to higher taxonomic levels if they are redundant or uninformative. In a test of six published microbiome studies, applying taxaHFE resulted in an 88% reduction in the number of features. Moreover, this feature reduction came at no cost to the downstream predictive power of the microbiome in machine learning models. Indeed, machine learning models using data processed with taxaHFE performed better than models trained on any single taxonomic level.
Beyond the microbiome, taxaHFE can be applied to other data structures containing hierarchical features. Dr. Mary Kable, a colleague of Dr. Lemay and Dr. Oliver, has investigated diet-microbiome relationships by placing foods on a “food tree” (PMID: 34958387). Closely related foods exist more proximally on the food tree, much like a phylogeny of microorganisms. taxaHFE can be applied to this novel method for diet analysis, allowing nutritionists to investigate which foods or general food groups are associated with a particular health outcome.
To develop taxaHFE, and the many other questions Dr. Oliver and Dr. Lemay investigate, access to HPC environments is critical. taxaHFE runs hundreds of models to collapse high dimensional data and was designed to utilize parallel compute resources. Containerization of taxaHFE, using Singularity, and the high performance of SCINet resources such as Ceres enables access to this method across ARS.
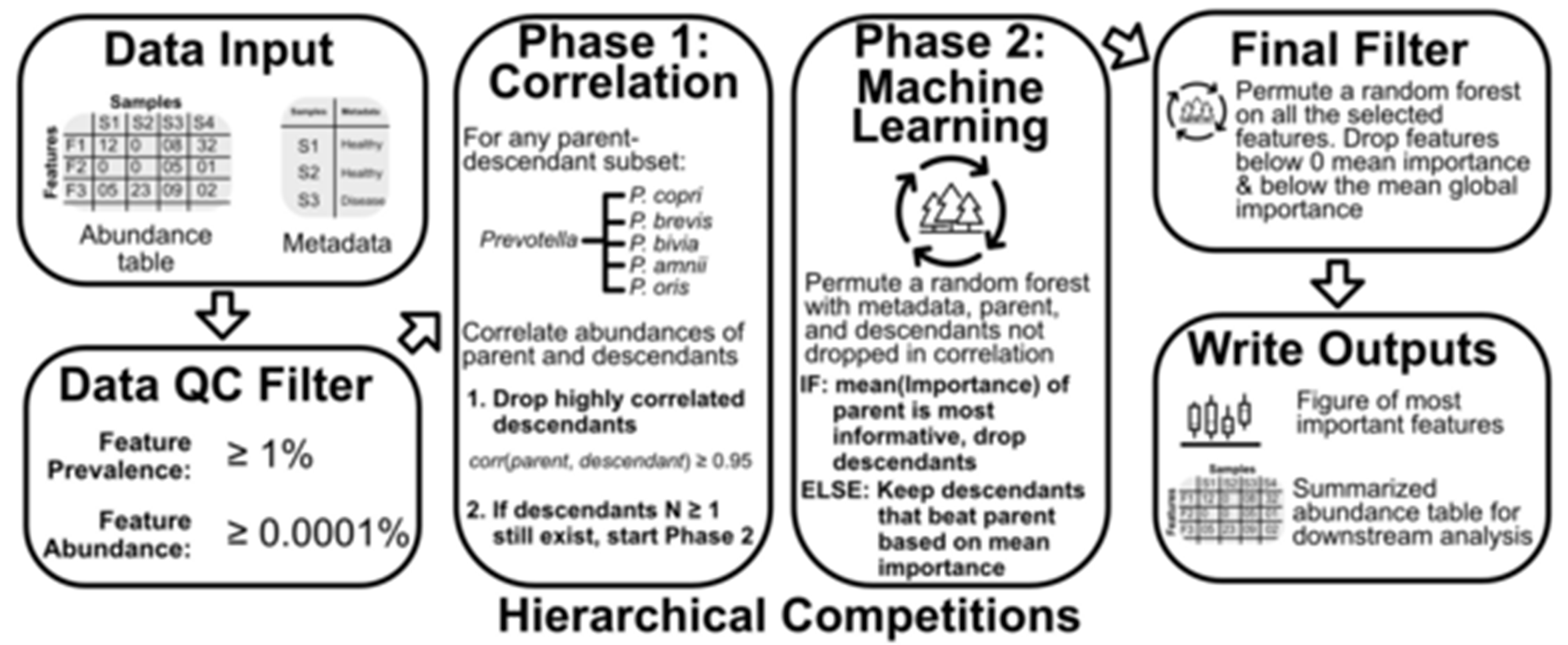
Figure 2: Outline of taxaHFE algorithm

