By: George Liu, Research Biologist (Bioinformatics) and Zhenbin Hu, Research Computational Biologist/ SCINet Postdoctoral Fellow | 07/31/2023 Animal Genomics and Improvement Laboratory, BARC, ARS, USDA, Beltsville, MD
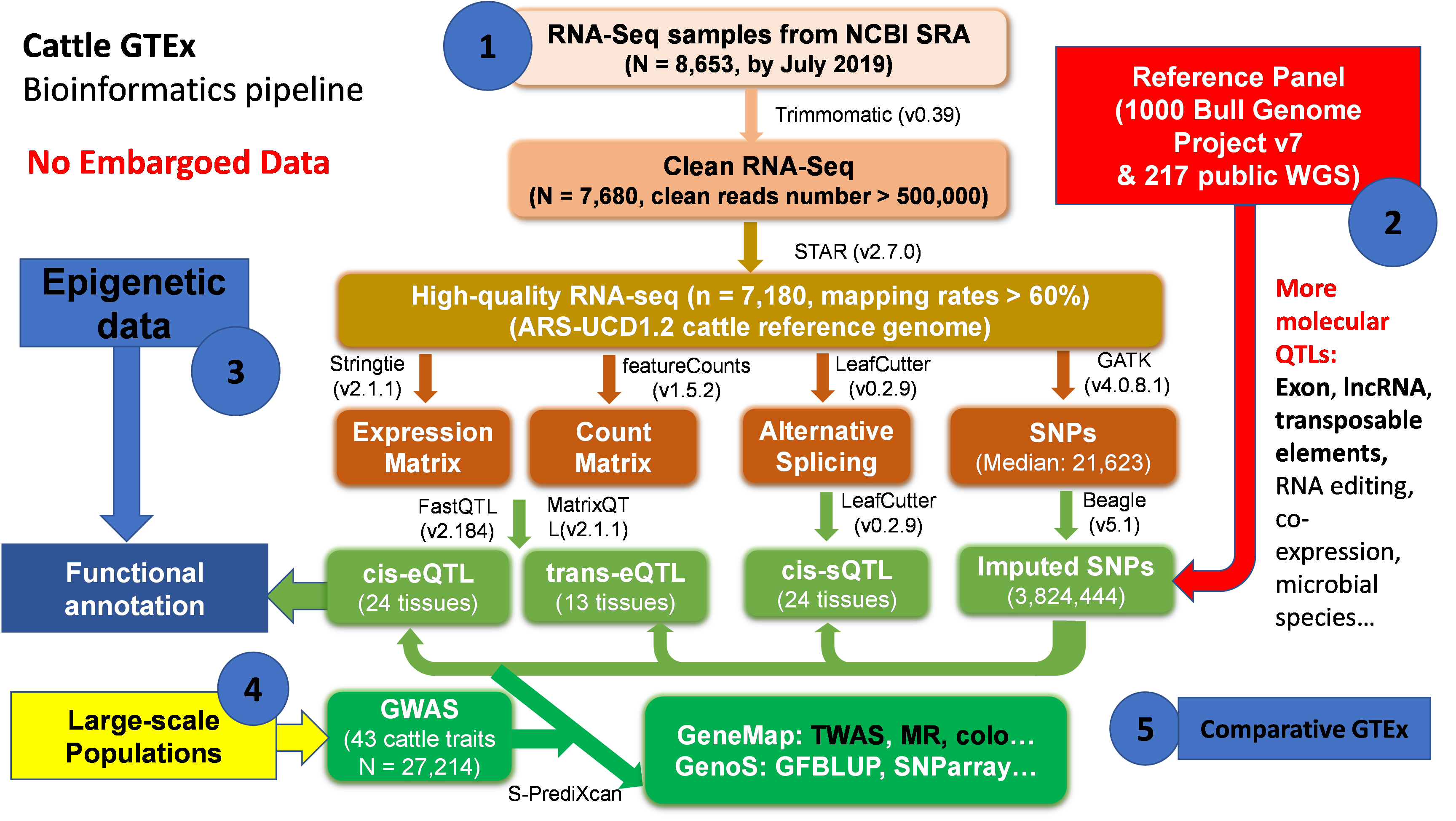 Figure 1. An overview of FarmGTEx-cattle data analysis and mining framework detailed in A multi-tissue atlas of regulatory variants in cattle. Nature Genetics 2022
Figure 1. An overview of FarmGTEx-cattle data analysis and mining framework detailed in A multi-tissue atlas of regulatory variants in cattle. Nature Genetics 2022
The Animal Genomics and Improvement Laboratory (AGIL) has a primary goal of discovering and developing improved methods for genetic and genomic evaluation of economically important traits in dairy animals and small ruminants. It also conducts fundamental genomics-based research to enhance animal health and productive efficiency. Dr. George Liu and his team have been diligently building comprehensive data resources and providing data analysis and mining tools for translational omics research.
Dr. Liu is also a co-founder of the Farm Animal Genotype-Tissue Expression (FarmGTEx) Consortium, which aims to create a comprehensive public resource for studying tissue-specific gene expression and regulation in major livestock species, including cattle, pigs, sheep, goats, and chickens. Since its official launch, the FarmGTEx Consortium has garnered interest from nearly 400 researchers in 48 countries. As a part of FarmGTEx, Dr. Liu co-led the development of the FarmGTEx databases for cattle, pigs, and chickens, with the CattleGTEx paper featured as a Nature Genetics cover story last year. FarmGTEx-related research has produced complete, open-access catalogs of regulatory elements for cattle, pigs, and chickens which are available on a public portal (https://www.farmgtex.org/), providing immense utility to the livestock community and industry. These resources collectively serve as primary references for animal genomics, breeding, adaptive evolution, comparative genomics, and veterinary medicine.
A critical factor contributing to our success has been the utilization of SCINet’s high-performance computing clusters (HPCs), including Ceres and Atlas. From the very beginning, SCINet has been an integral part of our large-scale data-driven research, enabling the processing of over 500 TB of omics data for mining. Our analyses have encompassed more than 5,000 whole-genome sequences and over 40,000 transcriptome datasets, yielding a wealth of genetic information critical for animal improvement. Additionally, SCINet resources facilitated the development of the Cattle Gene Atlas and enabled the first-ever transcriptome comparison between humans and cattle.
SCINet also provides us with a computing environment that empowers us to develop new data analysis tools using artificial intelligence (AI) and machine learning (ML). Dr. Zhenbin Hu, a SCINet/AI-COE Postdoctoral Fellow working alongside Dr. Liu, is currently employing ML to train a Convolutional Neural Network (CNN)-based model to construct high-quality databases of structural variations. He plans to customize and apply the Sparse Conditional Gaussian Graphical Model and other frameworks to the CattleGTEx data. Furthermore, Dr. Hu is also actively processing large-scale genomics data for the Sheep/GoatGTEx project as a core member. Looking ahead, we are confident that the large, distributed computing and GPU resources of SCINet will be essential for developing new pipelines, enabling more efficient execution of massive and complex data analysis jobs.
Dr. Liu and Dr. Hu are pleased to announce the formation of a new SCINet working group, the Translational Omics Working Group, to help facilitate omics-related research in ARS. Please see the working group page for more information about this working group, including how to join!

