By: Ben Rosen | 01/10/2021 USDA-ARS, Animal Genomics and Improvement Laboratory, Beltsville, MD
As a scientist in USDA-ARS’s Animal Genomics and Improvement Lab, one of our missions is to generate scientific resources used to improve our ability to raise cattle, sheep and goats to their fullest potential. We have sequencing technologies that can be used to sequence the DNA of these animals and generate a blueprint of their genomes. A genome consists of long strings of DNA called chromosomes. The problem we face is that the sequencing technologies can’t sequence full chromosomes, but rather they sequence millions of pieces of DNA or fragments of the chromosome that have to be pieced together like a puzzle using HPCs (high performance computer [HPC] clusters).
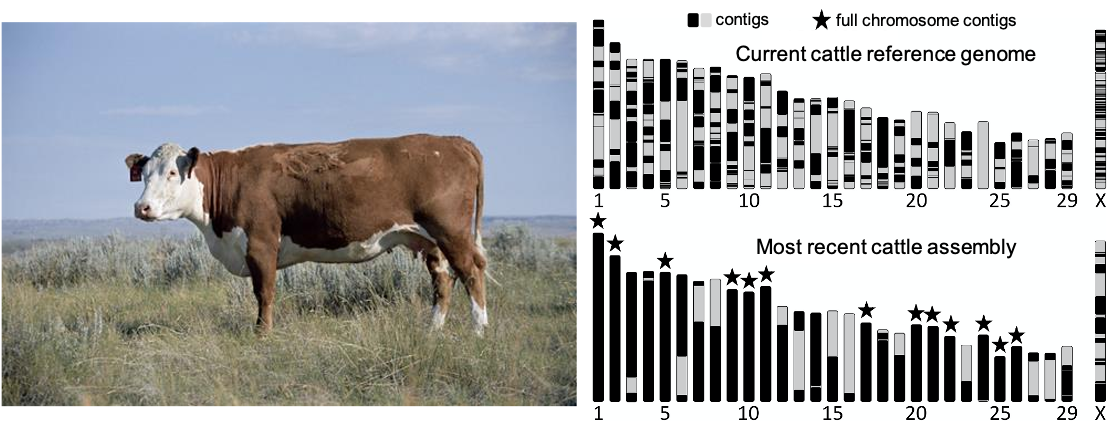
My main focus is to improve the accuracy and completeness of reference genome that are then used to, for example, search for disease-causing mutations or breed for a beneficial trait like milk production. Our most recent cattle genome that we pieced together had more unbroken chromosomes compared to the more highly fragmented (contigs) previous reference genome for cattle (see above image; Rosen et al. 2020, GigaScience). This more complete genome might have DNA that was missed in earlier assembled genomes and is important to researchers studying large-scale chromosome biology and its role in gene regulation. For example, we know that different breeds possess novel sequences and variation in how the chromosomes are arranged that are difficult or impossible to detect in a single reference genome based upon a single breed (Low et al. 2020, Nature Communications). Multiple assembled genomes are expanding the range of genetic diversity we are able to interrogate. Our ultimate goal is to incorporate all known genetic diversity into a single reference for each species called a pan-genome. This will require significant super computing resources that are only possible thanks to SCINet.
SCINet note: Not all genomes are assembled equally. Genome size and complexity have an enormous impact on the computational time required for assembly. The HPCs available through SCINet, named Ceres and Atlas, allow our research group to put together the pieces of DNA using different parameters of the program we use to determine the best possible solution. SCINet is a shared computing resource where users can purchase access to the HPC, but are able to use way more than they could afford for short periods of time. This approach has enabled us to triple our compute capacity without increasing the burden on cost or local IT resources. SCINet also has amazing staff that support the machines, software installation and domain experts as part of the Virtual Research Support Core (VRSC). This in-house ARS capacity is essential because we can now accomplish objectives that would never have been possible before.

