By: Yong-qiang Charles An | 01/10/2022 USDA-ARS Plant Genetics Research Unit at Danforth Plant Science Center, Saint Louis, MO
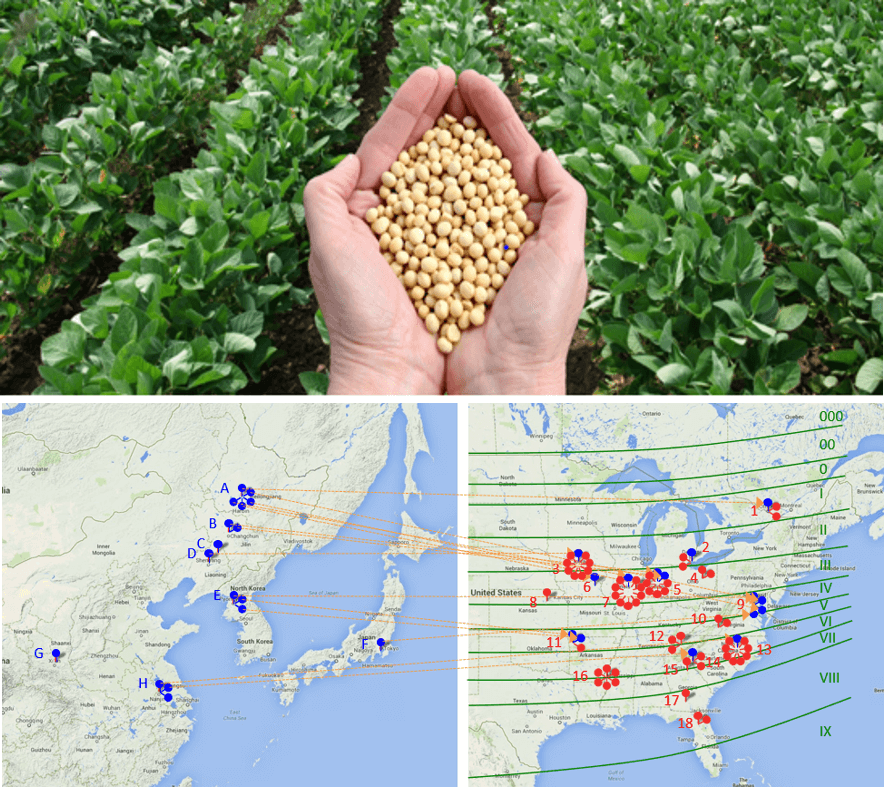
Photo: We sequenced and analyzed seed transcriptomes of 75 soybean lines representing 90 years of North American soybean breeding history for identifying breeder-selected genes and alleles. Ancestral soybeans (blue dots) of Asia were introduced in North America and used to breed the modern soybean cultivars (red dots) in North America
Soybean (Glycine max (L.) Merr.) is a versatile, nutrients-laden, economically invaluable crop with capacity to restore soil fertility through atmospheric nitrogen fixation. It holds great significance in ensuring adequate global nutritional food security and environment-friendly sustainable agriculture. With advances in next-generation sequencing and many other high-throughput technologies, worldwide soybean researchers have generated and made available a massive amount of complex multi-disciplinary datasets. These datasets provide an unprecedented opportunity to discover new traits and their causative genes and alleles, infer gene networks and build effective phenotypic prediction models that are critical for modern soybean improvement. My laboratory has been working on establishing a big-data driven, interdisciplinary technology platform by consolidating the in-house generated with open-access, diverse and massive amount of data. We have been developing a suite of data analysis, mining tools and strategies for translating the continuously increasing large datasets into improved soybean cultivars.
SCINet Note: SCINet, since its inception, has been an integral part of our large-scale data driven research. Overall, SCINet has enabled us to effectively produce more than 50 TB of soybean -omics data for mining. Using SCINet’s high performance computing clusters (HPCs; Ceres and/or Atlas), we consolidated and analyzed over 5,000 whole-genome sequences and 3,920 transcriptomes of soybean, which were generated in our laboratory or were available in the public domain. This venture resulted in a large-scale, expandable, and user-friendly genomic resource constituting structurally and functionally annotated 32 million single nucleotide polymorphisms (SNPs) and 3.3 million DNA structural variants (SVs) in 1,562 diverse soybean accessions. The transcriptome analyses delineated transcript accumulation of 56,000 soybean genes from 1,438 distinct biological treatments. Collectively, these datasets are of immense utility to research community and thus, a complete annotated SNPs dataset has been released in Ag Data Commons (An et al. 2020) and Soybase. A comprehensive description of the dataset and its versatile use will be published (Zhang et al. under review).
SCINet has provided access to pre-installed tools and software along with the capacity to download and install our own user-specific tools, which enables us to effectively develop a suite of new data analysis pipelines/ data-mining strategies. SCINet was also instrumental in our discovery of the genes underlying two large-effect protein and oil QTLs, which soybean researchers attempted to clone in past three decades, and demonstrating their key roles in soybean domestication (Zhang, et al. 2020).
References:
An Y-qC, Zhang H, Meryer R. 2020. Data from: Development of a versatile resource from 1500 diverse genomes for post-genomics research. Ag Data Commons. doi:10.15482/USDA.ADC/1519167
Zhang H, Goettel W, Song Q, Jiang H, Hu Z, Wang ML, An Y-qC. 2020. Selection of GmSWEET39 for oil and protein improvement in soybean. PLOS Genetics 16(11): e1009114. doi:10.1371/journal.pgen.1009114
Zhang H, Jiang H, Hu Z, Song Q, An Y. Accepted with monir revisions. Development of a versatile resource from 1500 diverse genomes for post-genomics research. BMC Genomic. bioRxiv: 2020.2011.2016.383950.

