By: Adam R. Rivers | 03/31/2022 USDA-ARS Genomics and Bioinformatics Research Unit - Gainesville
CRISPR-Cas is a powerful tool for gene editing. CRISPR can also be used to discover gene function by disrupting every gene in the genome and screening millions of cells in parallel. This “CRISPR screen” or “CRISPR pool” technique knocks out (disrupts) the function of thousands of genes in a large pool of cells. Each cell has only a single gene knockout, but the cells collectively have nearly every gene in their genome disrupted. This can be done in any bacterial culture or eukaryotic cell line where a vector can be introduced and cells can be grown in culture. The population of mutated cells is then grown in the presence of a compound like a toxin, drug, or metabolite. After growth, we extract DNA from control and treatment cell populations at the beginning and end of the experiment. The Cas cut sites are amplified and sequenced. These data are used to create a tally of the mutations in the control and treatment populations. Mutations that improve fitness in the presence of the compound are enriched and can be identified statistically. At the end of an experiment, we have a small list of genes that are likely involved in responding to the compound. These genes can be investigated in more detail to work out the exact mechanisms of their effect.
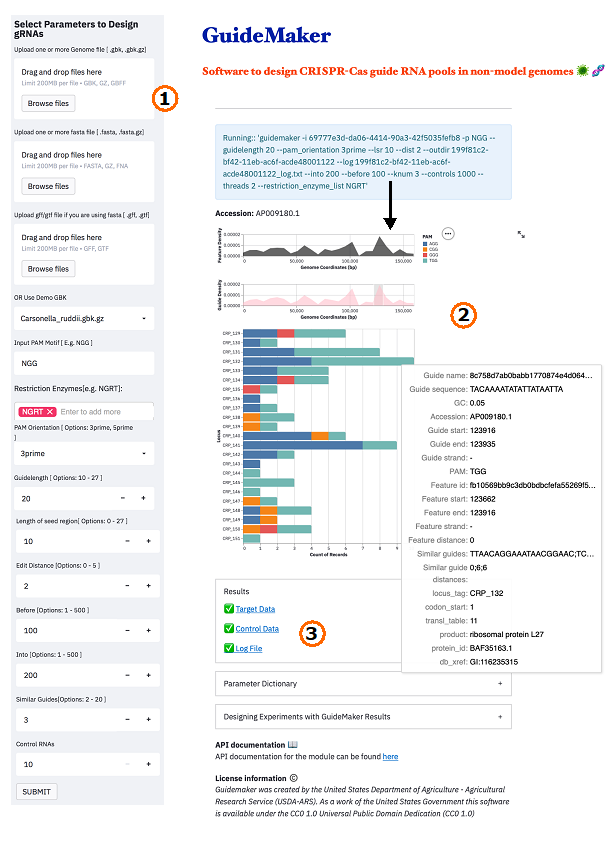
Cas enzymes in a CRISPR-Cas system target a specific site in the genome for cleavage by recognizing a Protospacer Adjacent Motif (PAM) site and an adjacent target site in the genome. Each Cas enzyme type recognizes its own PAM sites, for example, SpCas9 recognizes sites in the genome with the nucleotides “NGG”. Cas cleavage is site-specific because it only cuts in the presence of a guide RNA sequence that has a specific ~20-25 bp long sequencing matching a point in the genome next to a PAM site. Designing these guides to target every gene in the genome multiple times is challenging. Every guide RNA needs to target a single site in the genome and needs to be somewhat different from similar guides for specificity. There are also design considerations for how efficient a guide will be at knocking out a gene. Once the complicated design process is complete, the guide oligonucleotides can be commercially synthesized in one large pool. The pooled guide oligonucleotides are then cloned, amplified, and put into an expression vector that allows it to be expressed in a population of cells. There are standard molecular tools to do this in most organisms.
One of the major challenges of conducting CRISPR screens is designing the guide RNAs that target specific sites in the genome. This is especially true when dealing with a non-model organism or a less common Cas enzyme system. To address this problem, my group, in collaboration with researchers at the University of Florida, created user-friendly guide design software that can rapidly create guides for any genome or Cas enzyme. A web application version of GuideMaker is hosted at https://guidemaker.app.scinet.usda.gov and as a tool in the Cyverse Discovery Environment. A command-line version of the software is also available at https://guidemaker.org. Under the hood, GuideMaker implements several unique machine learning methods including a powerful method for approximate nearest neighbor search (HNSW) and a model to predict on-target binding originally designed by Microsoft Research. I hope that my software can bring the power of CRISPR functional gene discovery to more scientists at ARS and I am happy to help ARS scientists get started with the method. SCINet resources were critical to the development of the software. We extensively benchmarked the software using multiple compute nodes on Ceres and Atlas. An article describing the software in detail is in press at the journal GigaScience.