By: Michael A. Alcorn | 01/20/2023 AI Center of Excellence Fellow
Over the past 20 years, the Western United States has experienced several widespread outbreaks of vesicular stomatitis (VS), a disease that affects livestock. Although infected animals generally clear VS without serious long-term effects, the initial symptoms (e.g., blisters on the lips) resemble foot-and-mouth disease (FMD), which is a much more severe illness that can wreak havoc on the agriculture sector of the economy. As a result, developing methods for determining as early as possible whether VS/FMD-like symptoms are actually VS or FMD is of pressing importance. One potentially informative signal for assessing the likelihood of a VS infection is the likelihood that the disease is spreading in nearby regions.
AI Center of Excellence Fellow Dr. Michael Alcorn has been researching the potential for using spatiotemporal deep neural networks (STNNs) to model VS risk through both space and time. Specifically, he has investigated two alternative approaches for encoding spatial inputs—grids and graphs—and two corresponding STNNs—convolutional long short-term memory recurrent neural networks (ConvLSTMs) and graph ConvLSTMs (GConvLSTMs). To train the STNNs, Dr. Alcorn worked with former SCINet Fellow Dr. Kerrie Geil and collaborators to construct the largest dataset so far of geotagged case reports for VS in the U.S. and Mexico from 2001 to 2020. These case reports were then linked to temporally varying environmental and anthropogenic variables recorded at a 30 km2 resolution on a national scale, all of which was accomplished using SCINet’s high-performance computing clusters.
When modeling reported VS cases in the U.S. spatially, the ConvLSTM struggled—likely because of the rarity of reported VS cases in the dataset; however, the ConvLSTM was more effective than a non-STNN and linear model when modeling the aggregated VS cases per month. In contrast, the GConvLSTM was more effective than a non-STNN and linear model when modeling reported VS cases in the U.S. county-wise, and it identified plausible regions of VS risk. Additionally, when jointly trained on U.S. and Mexico data, the GConvLSTM’s performance on U.S. counties slightly improved, which suggests there are shared factors influencing VS spread between the two countries. Together, these results (a) highlight the importance of accounting for spatiotemporal dynamics when modeling the spread of livestock diseases and (b) lay a foundation for further exploration of STNNs as models for investigating these systems.
The code for implementing and training the STNNs was written in Python and uses the deep learning library PyTorch. The code for the project will be released as an open-source GitHub repository soon, which will allow other scientists to easily adapt these state-of-theart machine learning techniques to their own projects. This work was presented at the 2022 AGU Fall Meeting, and slides for that presentation can be found here.
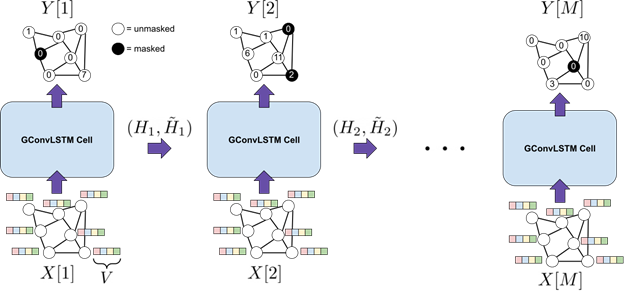
Figure 1. An overview of the graph spatiotemporal deep neural network that was trained to model the spread of vesicular stomatitis. The input to the model is a sequence of graphs where each node contains environmental and anthropogenic variables for a specific county in a specific month. The output of the model is also a sequence of graphs where each node indicates the reported number of vesicular stomatitis cases for a specific county in a specific month.

