By: John Humphreys | 07/16/2020 USDA-ARS, Range Management Research Unit, Las Cruces, NM
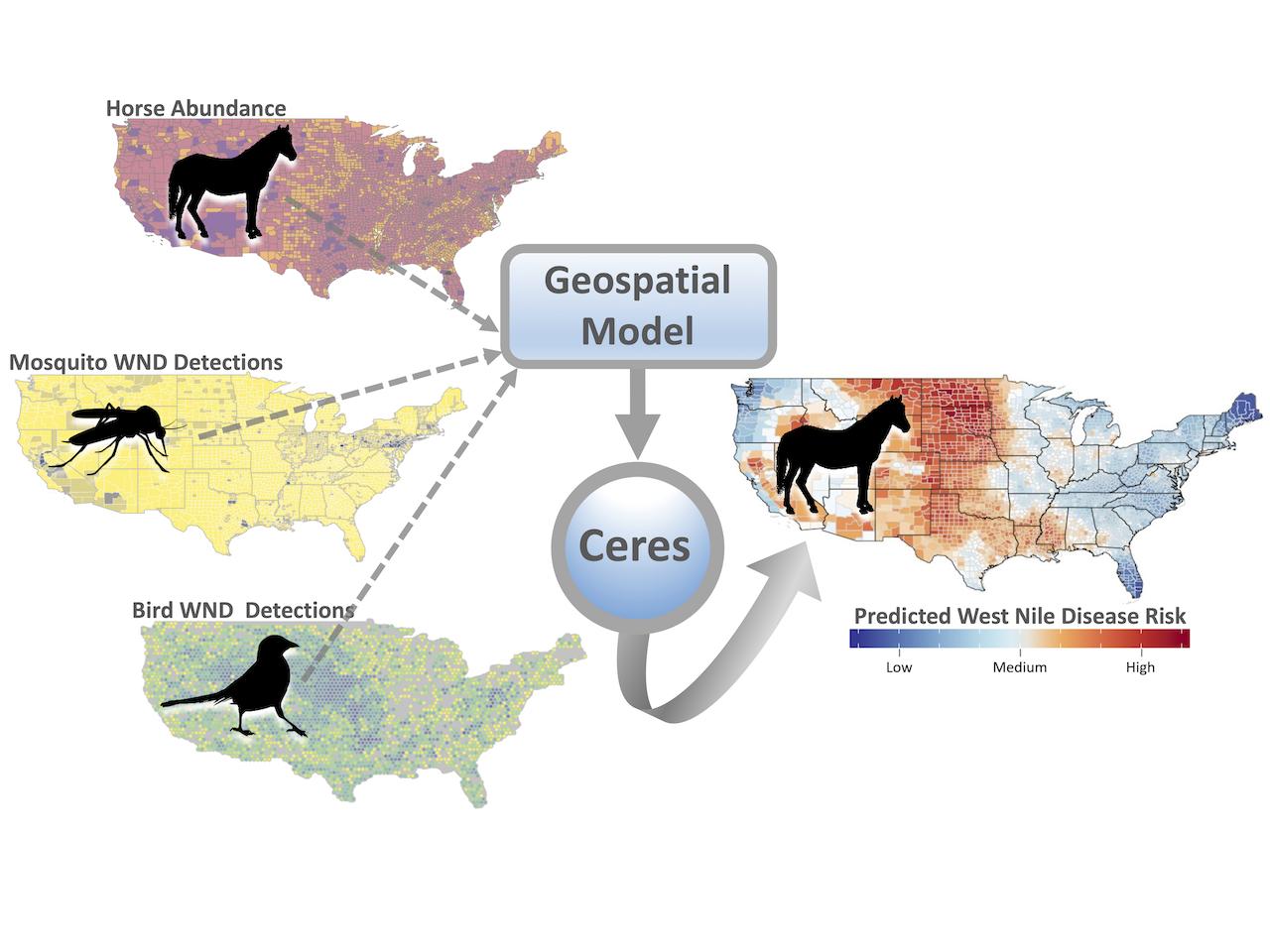
A fundamental need to meet USDA ARS’s Grand Challenge initiative is to improve agricultural production while reducing the impact of the emerging pests, pathogens, and invasive species that threaten US livestock. Pathogens such as West Nile Virus (WNV), Vesicular Stomatitis Virus (VSV), and others impair livestock health, deplete veterinary resources, and threaten agricultural trade. To better anticipate and prepare for future disease outbreaks caused by viruses, it is essential to model the virus-vector-host interactions and environmental factors that drive disease spread across geographic space and through time. Because disease models must provide high-resolution outputs across expansive geographic extents while simultaneously accounting for the correlations that exist in input variables in both the temporal and spatial dimensions, analyses are often too computationally demanding for traditional hardware and necessitate use of high-performance computing resources like those available through SCINet.
As part of the ARS Predictive Disease Ecology Grand Challenge Project led by Drs. Deb Peters and Luis Rodriguez, a spatiotemporal disease model was developed to forecast future West Nile Disease (WND) outbreaks in horses across the continental US. Postdoc Dr. John Humphreys led the analysis to predict the distribution and timing of future WND outbreaks. The Centers for Disease Control and Prevention (CDC) records provided the count of veterinary-reported WND cases for horses between 2000 – 2018 aggregated by county. The team used the USDA National Agricultural Statistics Service database to map horse populations, incorporated CDC mosquito surveillance reports to identify insect vector ranges, and analyzed more than 10 million bird occurrence records from the Cornell Laboratory of Ornithology to map the distributions of avian species known to host WNV. These datasets allowed the model to link the at-risk livestock population (horses) to times and locations with both the WNV reservoirs (birds) and the WNV vectors (mosquitos) that transmit the virus between those reservoirs and livestock. The research team applied a Bayesian hierarchical modeling framework to construct the model and specified that the prediction for any one location be dependent on the disease risk estimated for surrounding areas and past times (manuscript is in preparation).
SCINet Note: After 24 hours of processing on a laptop (Intel Core Processor i9-8950HK, 8 Core, 2.9GHz), the model was only approximately 20% completed, with multiple model versions to be run. Conversely, running the model on the Ceres HPC completed in less than 10 hours, dramatically reducing the processing time and freeing up the laptop for other uses. Preparation for running the model on Ceres entailed uploading the existing R script and model input data (using Globus), creating a text file specifying the number of nodes, cores, and memory needed, and then submitting that file to the Ceres job scheduler and management system (Slurm). The team opted to double what was available on their laptop and requested two nodes each with 8 cores. Conveniently, the R package used to run the model (r-INLA) included native multithreading (OpenMP) to handle parallel processing and allowed HPC cores to run concurrently.

