Research Spotlight
Comparative genomics reveals a light-activated phytotoxin that contributes to red leaf blotch disease of soybean
Nicholas Greatens1,2 and Rachel A. Koch Bach2
1SCINet Program and ARS AI Center of Excellence, Office of National Programs, Agricultural Research Service, United States Department of Agriculture, Beltsville, Maryland, United States of America
2Foreign Disease-Weed Science Research Unit, Agricultural Research Service, United States Department of Agriculture, Fort Detrick, Maryland, United States of America
Coniothyrium glycines causes red leaf blotch, a major disease of soybean in Africa (Figure 1). It is one of two fungal pathogens listed on the USDA APHIS Plant Protection and Quarantine Select Agents and Toxins list owing to its likely destructive potential if it spreads to major soybean growing regions.
At the USDA-ARS Foreign Disease-Weed Science Research Unit, based in Fort Detrick, Maryland, SCINet/AI-COE Postdoctoral Fellow Nicholas Greatens and Research Plant Pathologist Rachel Koch Bach study this carefully regulated pathogen in containment research facilities. Drawing from recent collections of the red leaf blotch pathogen from across southern and eastern Africa, ARS researchers have published the first annotated genome of C. glycines and provided new insights into how it causes disease in soybean. Genome assembly and annotation and comparative genomics analyses were made possible by SCINet’s Ceres supercomputer and the personnel and educational resources of SCINet.
Many leaf spot fungi produce compounds that are toxic to plants, and these phytotoxins can play key roles in causing disease. Often, the enzymes that synthesize phytotoxins are encoded by closely linked genes that act together and are inherited as a cluster. This close linkage enables detection of these gene clusters and helps to predict their chemical products.
Examination of the C. glycines genome with antiSMASH software revealed a gene cluster similar in structure to well-studied clusters that produce cercosporin and elsinochrome, light-activated, disease-causing toxins produced by pathogens like Cercospora beticola, causing Cercospora leaf spot of beet, and Parastagonospora nodorum, causing Septoria leaf blotch of wheat. The red leaf blotch symptom caused by C. glycines resembles the distinct reddish leaf spots of Cercospora diseases.
In cultures of C. glycines grown under light conditions, the gene cluster was upregulated, and in leaves inoculated under light, more lesions developed and at a faster rate. Liquid chromatography and mass spectrometry confirmed production of elsinochrome in field-collected specimens and pure cultures.
Remarkably, similar gene clusters likely synthesizing elsinochrome or related compounds were detected for the first time in other important plant pathogens and fungi using BLAST, phylogenetic approaches, and the software CAGECAT, a user-friendly tool useful for exploring biosynthetic gene clusters. As it turns out, elsinochrome and related compounds may have broader importance to fungal ecology than was previously understood.
This study lays the groundwork for understanding the molecular machinery required for plant pathogenicity by the Select Agent C. glycines. Understanding the molecular basis of disease is an important step toward the development of effective management strategies for red leaf blotch of soybean. The study is in press at PLOS One as “Production of the light-activated elsinochrome phytotoxin in the soybean pathogen Coniothyrium glycines hints at virulence factor”.

Figure 1. Red leaf blotch disease in soybean.
News
An improved queue/partition scheme for Ceres
For a variety of reasons, Ceres has accumulated many queues/partitions over the years, now numbering well over a dozen. Having so many partitions can make Ceres challenging to use, especially for new users. (If you are not sure what “partition” or “queue” means on a supercomputer, check out our user guides for more information!) During Ceres’ February 2025 maintenance, we added a new partition, named “ceres”, that greatly simplifies running compute jobs on Ceres. The bottom line is that you should now be able to submit nearly all jobs to the “ceres” partition and effectively ignore the remaining partitions. In the future, some or all of these “legacy” partitions will be removed.
The new “ceres” partition includes all community (that is, non-priority) nodes, and we expect that use of this new partition will ultimately result in an improved user experience, shorter wait times, better cluster utilization, and a more similar user experience on Ceres and Atlas. Please note, however, that the new “ceres” partition has a default job time of 2 hours. This will help avoid long wait times in the queue due to implicitly requesting far more time than a job needs, but it also means that if your job requires more than 2 hours, you will need to explicitly request more time. Please see the SCINet website for more information.
New image annotation software on Atlas
You can now easily create computer vision model training datasets directly on Atlas without needing to transfer image files to and from local laptops or workstations for annotation. Labelme, an image annotation application, is now available as an interactive app on Open OnDemand on Atlas. Labelme supports a variety of annotation types, including bounding boxes and custom polygons, and it also supports automated and semi-automated annotation using pretrained, general computer vision AI models.
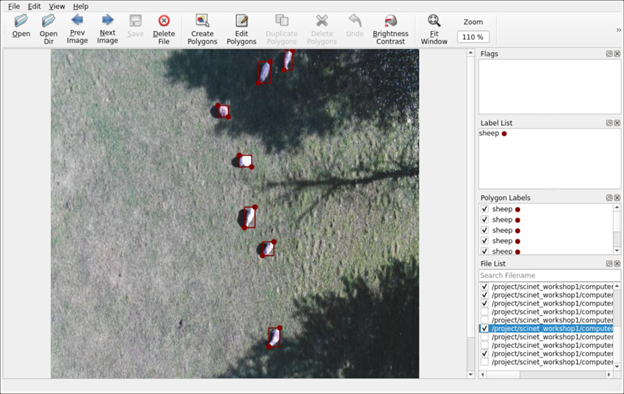
Figure 2. Labelme running on Atlas.
Would you like to help test SCINet’s new GPU nodes?
Usage of SCINet’s GPU resources has increased dramatically over the past few years, and SCINet is expanding its GPU offerings to ensure we can continue to meet demand. We’ve recently added 12 new GPU nodes to Atlas, each with 4 NVIDIA L40S GPUs. We are looking for SCINet users to help us test these new GPU nodes before they go into full production! We’ll ask early testers to try running their usual GPU workloads on the new nodes and provide feedback about their experience. If you’d like to help, please contact Brian Stucky (brian.stucky@usda.gov) or Heather Savoy (heather.savoy@usda.gov), and thank you!
AI-COE/SCINet internships update
We are pleased to announce that we have matched 15 graduate student interns with ARS AI-COE/SCINet internship opportunities based on the mutual interests of each student and their prospective mentor. These interns will work on a wide variety of research projects, all of which include significant artificial intelligence/machine learning or data science components. Twelve of these internships will begin this summer; three interns are already working with ARS mentors in spring internships. Many thanks to the 26 ARS scientists who applied to serve as AI-COE/SCINet graduate student internship mentors in 2025!
We are again planning a virtual internships research symposium in the fall, and we expect to announce details this summer!
AI Innovation Fund and SCINet/AI-COE postdoctoral fellowship mentor proposals update
If you submitted a proposal last December for an AI Innovation Fund award or to serve as a SCINet/AI-COE postdoctoral fellowship mentor, you have undoubtedly been wondering about the status of your proposal. We are pausing both of these programs at this time. We plan to re-evaluate later this year and will provide additional updates if we are able to move forward with either program.
Working Group Updates
SCINet working groups (WGs) support ARS researchers and their collaborators in using scientific computing methods and SCINet computational resources in their research. Common WG activities include hosting recurring virtual meetings and webinars, organizing training events, and participating in collaborative research or software development projects.
Translational Omics Working Group
The Translational Omics Working Group is hosting an upcoming webinar:
Improving Production Traits in Poultry through Genome Editing
- Thursday, May 8, 2025, 1 PM ET
- Paula Chen, Ph.D., U.S. Department of Agriculture, Agriculture Research Service, Plant Genetics Research Unit, Columbia, MO 65211
For more information or to join the working group’s email list, please contact George Liu (George.Liu@usda.gov) or Wenli Li (Wenli.Li@usda.gov)
Protein Function and Phenotype Prediction Working Group
Protein structure prediction resources continue to expand on the SCINet clusters!
AlphaFold 3 is now available on both the Ceres and Atlas clusters! Special thanks to the SCINet Virtual Research Support Core for making this possible. Instructions for running AlphaFold 3 on SCINet can be found in the SCINet guide.
In addition to AlphaFold 3, SCINet has also introduced user-friendly modules for several other leading protein structure prediction tools, including AlphaFold 2, ESMFold, and OmegaFold. Training materials for running AlphaFold 2, ESMFold, and OmegaFold on SCINet are available here. Additional tools, such as Boltz-1, have been successfully deployed on the Atlas GPU nodes.
Our working group is benchmarking these tools, and the results will be published soon. The figure below shows a preview of the comparisons between AlphaFold 2 and AlphaFold 3 on a maize protein dataset. Early findings suggest that AlphaFold 3 makes predictions substantially faster, effectively handles diverse biomolecular complexes (proteins, DNA, and ligands), and has only a slight reduction in prediction accuracy compared to the previous version.
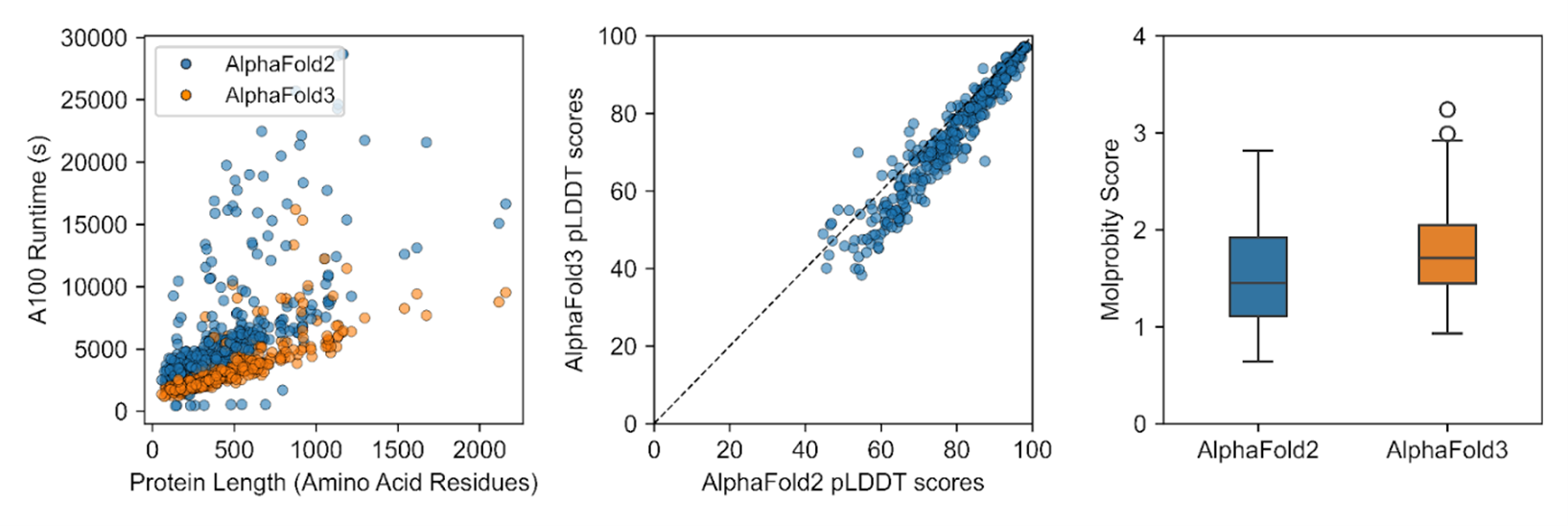
Figure 3. Left: Runtimes for generating structures of 417 maize genes using one Atlas A100 GPU. Middle: Pairwise predicted Local Distance Difference Test (pLDDT) scores, showing AlphaFold 2 models generally have higher confidence scores. Right: MolProbity scores indicate structural quality was similar between AlphaFold 2 and AlphaFold 3 predictions.
Geospatial Research Working Group
Several Geospatial Research Working Group members have leveraged SCINet resources in recent manuscripts:
- Dr. Georgia Harrison led a team of USDA, USGS, and BLM researchers in assessing the accuracy of the Rangeland Analysis Platform’s satellite-derived fractional cover estimates with out-of-sample field sites in Ecological Indicators https://doi.org/10.1016/j.ecolind.2025.113267.
- Drs. Alexander Hernandez and Efrain Duarte and team leveraged UAVs to predict soil moisture in semiarid ecosystems across the western US in Geocarto International: https://doi.org/10.1080/10106049.2025.2461523. Dr. Duarte is a current SCINet/AI-COE fellow.
Animal Behavior AI Working Group
This SCINet working group aims to explore the potential benefits of Artificial Intelligence (AI) in animal behavior research. The working group will provide an open platform where participants can share knowledge, discuss challenges, and explore opportunities for leveraging high-performance computing resources through SCINet to advance animal behavior research. By leveraging cutting-edge AI techniques, we aim to tackle the potential limitations of traditional methods, including challenges with handling large datasets and the computational demands of advanced analyses.
- Please sign up for the working group
- Contact: John Y. Park, SCINet/AI-COE Postdoctoral Fellow. (john.park3@usda.gov)
- Working group page
- Our team on Teams
Training
Training workshops
Foundations in bioinformatics
Lead(s): Genome Informatics Facility at Iowa State University and SCINet Office
Starting in April, the SCINet Office is offering a series of workshops to help ARS researchers develop practical skills for using bioinformatics in their research. The workshops in this series are designed to provide a thorough introduction to modern bioinformatics concepts, techniques, and best practices. Some of the workshops in the series were originally developed for, and offered at, the Forum on AI Applications to USDA Science in College Station, TX in 2024. We are offering them as virtual workshops in 2025, with expanded content and more opportunities for hands-on practice.
Series Outline:
- April 15, 2025, 1-4 PM ET: Introduction to modern bioinformatics
- April 17, 2025, 1-5 PM ET: Introduction to HPC environments and project management and organization
- April 29, 2025, 1-5 PM ET: Data preparation and quality assessment in genome assembly
- May 1, 2025, 1-5 PM ET: Genome assembly validation and improvement
- May 13, 2025, 1-5 PM ET: Introduction to RNA-seq analysis
- May 15, 2025, 1-5 PM ET: RNA for genome annotation and reproducibility in bioinformatics
At this time, registration is closed as we have reached maximum capacity for all workshops. However, you may complete the registration form to be added to our waitlist for future offerings.
From reads to variants: a pipeline for variant calling using DeepVariant
June 3, 2025, 1-5 PM ET
Lead(s): Sheina Sim (ARS Research Biologist), Craig Carlson (ARS Research Geneticist), and Haley Arnold (SCINet/AI-COE fellow)
DeepVariant is a DNA sequence variant caller that uses a convolutional neural network (CNN) to call genotypes relative to a reference genome assembly. In this workshop, we will discuss a workflow for calling variants from whole-genome data for multiple individuals. This workflow involves trimming and filtering raw reads, mapping them to a reference assembly, calling variants for each individual, merging the variants of all individuals into a single variant call format file (.vcf), and filtering the resulting variant file. We will guide participants through this pipeline step by step, providing generalized commands for each phase of the process, as well as strategies for optimizing cluster usage and reducing compute time. The final product will be a .vcf containing variants for all individuals which can be used for downstream analyses, along with a solid understanding of performing variant detection using DeepVariant.
If you did not have a chance to attend this workshop at the 2024 Forum on AI Applications to USDA Science, please join us!
To register, please complete this registration form.
The Carpentries instructor training
SCINet is collaborating with The Carpentries to offer The Carpentries’ Instructor Training Course for ARS scientists. In this course, you will learn about evidence-based practices for effective and inclusive teaching, with a particular focus on teaching computational skills. There is no fee charged to course participants, but seats are limited. If you are interested in becoming a Carpentries-certified instructor, please complete the instructor training form.
Coursera
The SCINet Office and the AI-COE are excited to provide training opportunities through Coursera. Coursera licenses are available to ARS scientists and support staff for training focused on scientific computing, data science, artificial intelligence, and related topics. Successful completion of courses and specializations result in widely recognized certificates and credentials.
Please visit the SCINet Coursera Training Page to request a license. Licenses will be assigned on a rolling basis and are active for three months. Users may be able to extend their licenses upon request.
Workshop reports
Machine learning (ML) and AI workshop series
Lead(s): SCINet Office
Our ML/AI workshop series wrapped up last month and consisted of three workshops:
- An Introduction to machine learning for science
- Computer vision I: introduction and image classification
- Computer vision II: object detection and instance segmentation
Throughout this series, participants learned how to train, evaluate, and use a variety of machine learning models for data analysis, including deep learning-based computer vision models for image classification, object detection, and instance segmentation. Due to the high interest in this series, we will be offering these workshops again. Click here to join our waitlist!
Transfer learning workshop
Lead(s): Research Computing team at the University of Florida
A transfer learning workshop, part of the Practicum AI series offered in collaboration with the Research Computing team at the University of Florida, was held on April 22 & 24, 2025. This course expanded on content from our AI/ML workshop series and helped participants learn different types of transfer learning techniques, such as feature extraction and fine-tuning. The course recordings and tutorial instructions are available on the SCINet website.
Please help us improve our training offerings!
What scientific computing training do you need? The SCINet Office’s goal is to provide training opportunities and resources that meet the needs of ARS researchers, so we would be grateful if you could complete our short training request form and let us know how we can best help you learn the computing skills you need. Your feedback will help us decide where we should focus our efforts over the next year and beyond.
Training opportunities are continually being updated on the SCINet Upcoming Events webpage. For more information on any of the above trainings, registration questions, or suggestions, please email SCINet-training@usda.gov.
Support
Getting Started with SCINet is as easy as 1,2,3
If you do not already have a SCINet account, we hope you will consider joining the 2,300+ researchers who do. Follow the steps below to get started with SCINet.

- Request a SCINet account to gain access to computational and training resources.
- Read the SCINet FAQs covering helpful topics such as account management, accessing and installing software, obtaining storage space for your project(s), and how to get technical help.
- Visit the SCINet Forum to connect to other users, ask questions, and learn how SCINet can enable your research. P.S. Don’t forget to complete your annual USDA information security awareness training! This is required to maintain your account. For technical assistance with your SCINet account, please email scinet_vrsc@usda.gov.
Support email addresses
All requests for help with user accounts, login problems, resource requests, or support for the Ceres HPC cluster should be sent to the SCINet Virtual Research Support Core (VRSC) at scinet_vrsc@usda.gov. Help requests specific to the Atlas HPC cluster should be sent to help-usda@hpc.msstate.edu.
Many emails are currently being sent to other SCINet email inboxes. For the most expedient response to your support requests, be sure to send them to scinet_vrsc@usda.gov or to help-usda@hpc.msstate.edu for Atlas-specific requests.
SCINet user tip
Do your analyses require or generate large amounts of data? Do you need long-term storage for your large datasets? SCINet strives to provide the storage that you and your workflows need to do your research, be it 1 TB, 20 TB, or more! If you are needing more working space on the clusters or more long-term storage space on Juno, you can request an increase for your existing project following the instructions here. If you are interested in a creating a SCINet project with new storage allocations to facilitate your research, see instructions here. Either way, we want to meet your data storage needs!
Do you have tips to share? Email them to ARS-SCINet-Office@usda.gov to be included in future newsletters.
SCINet Corner
SCINet Corner is a VRSC-moderated virtual space for people to share knowledge, discuss best practices, learn about new opportunities, and explore resources to support progress on their projects.
The next SCINet Corner will be held on May 22, 2025, from 1 – 2 PM ET. May’s event will focus on common Slurm parameters on both the command line and Open OnDemand, including using the new “ceres” partition on Ceres.
You can register for this and future SCINet Corners here.
Have a question that just can’t wait? Want to see what other users are doing? Reach out to the ever-expanding SCINet Forum community for ideas, support, or just someone to bounce ideas off of at https://forum.scinet.usda.gov/.
Connect
The SCINet Community
To see all the SCINet community updates and review past newsletters, visit the Newsletter Archive.
Contribute
Do you use SCINet for your research? We would love to share your story! Email ARS-SCINet-Office@usda.gov to contribute content, ask questions, or provide feedback on the SCINet newsletter or website.
SCINet Office
Haitao Huang, Computational Biologist
Moe Richert, Web Developer
Lavida Rogers, Training Coordinator
Heather Savoy, Computational Biologist
Brian Stucky, Computational Biologist, Acting Chief Scientific Information Officer
SCINet Leadership Team
Brian Stucky, Acting Chief Scientific Information Officer
Rob Butler, SCINet Program Manager
Jeremy Edwards, Science Advisory Committee (SAC) Chair
Jeff Silverstein, Associate Administrator

