Research Spotlight
Advancing Irrigation Mapping Through AI and Remote Sensing
S. Kossi Nouwakpo1 and Oishee Hoque2
1Northwest Irrigation and Soils Research Laboratory, Agricultural Research Service Kimberly, Idaho, United States of America
2Department of Computer Science, University of Virginia, Charlottesville, VA, United States of America
Efficient irrigation practices are critical to sustain profitable agriculture in many water-limited regions of the United States and beyond. However, irrigation types, whether sprinkler, surface, or drip irrigation, can vary widely in their efficiency and their influence on both water and soil resources. Accurate mapping of irrigation methods is essential for tackling water scarcity across large regions, boosting agricultural productivity, and informing conservation practices.
Researchers at the USDA-ARS Northwest Irrigation and Soils Research Laboratory at Kimberly, Idaho are spearheading the development of an irrigation methods mapping tool by leveraging Artificial Intelligence (AI) and publicly available satellite imagery. This project has greatly benefited from the contribution of Ms. Oishee Hoque, who was a 2023 ARS Artificial Intelligence Center of Excellence (AI-COE) / SCINet graduate student intern affiliated with the AI Institute for Next Generation Food Systems (AIFS). Ms. Hoque is a Ph.D. candidate in Computer Science at the University of Virginia whose expertise in AI enabled the creation of a large-scale dataset, the design of novel deep learning models, and the implementation of knowledge-informed frameworks for robust irrigation classification.
The team’s journey began with the development of IrrNet, the first deep learning model designed specifically for irrigation type mapping from satellite imagery. IrrNet tackles the difficulty of segmenting agricultural fields – especially when different irrigation types coexist in close proximity – by using a progressive patch size training strategy. The model starts with small image patches to capture fine-grained details and gradually increases patch size to generalize across broader spatial patterns. This simple yet effective approach improves semantic segmentation accuracy by up to 20% over standard models like U-Net and DeepLabv3+. IrrNet also demonstrated the importance of leveraging non-RGB spectral bands from Landsat imagery, such as near-infrared and thermal channels, for better irrigation-type differentiation.
Building on this momentum, the researchers introduced IrrMap, the first large-scale, ML-ready dataset tailored to irrigation method mapping (Figure 1). IrrMap includes over 1.1 million 224×224 patches extracted from Landsat and Sentinel imagery, with precise labels distinguishing sprinkler, flood, drip, and other irrigation methods. The dataset spans four U.S. states – Arizona, Colorado, Utah, and Washington – covering 14 million acres and 1.7 million farms. It integrates crop type, land use, and vegetation indices to provide rich contextual information. IrrMap includes supporting software and benchmark models that lower the barrier for others to build and evaluate irrigation classifiers.
To further improve generalization across regions and data modalities, the team developed KIIM (Knowledge-Informed Irrigation Mapping). KIIM combines vision transformer models for image segmentation with structured agronomic knowledge, such as crop-irrigation likelihoods . KIIM includes innovative model components that enhance the fusion of disparate RGB and vegetation index input data. KIIM achieves state-of-the-art performance, with over 70% intersection over union (IoU) improvement compared to a baseline Swin Transformer model for hard-to-classify drip irrigation in certain states, and requires only 40% of the training data to match baseline accuracy—demonstrating strong data efficiency.
Most recently, the team released IRRISIGHT, a multimodal dataset and scalable pipeline to support irrigation and water availability research. Covering 20 U.S. states and more than 1.4 million patches, IRRISIGHT integrates satellite data with soil, hydrology, evapotranspiration, and climate variables. It includes natural language prompts generated from soil properties, enabling experimentation with vision-language models for agricultural monitoring tasks. IRRISIGHT provides a new frontier for modeling irrigation decisions, drought resilience, and sustainable water use.
From IrrNet to IRRISIGHT, this body of work has significantly advanced the science of irrigation mapping. With openly released models, datasets, and code, it empowers the broader research community to tackle water sustainability at scale. More information can be found at the following links: IrrNet; IrrMap – paper, codes; KIIM – paper, codes; IRRISIGHT.
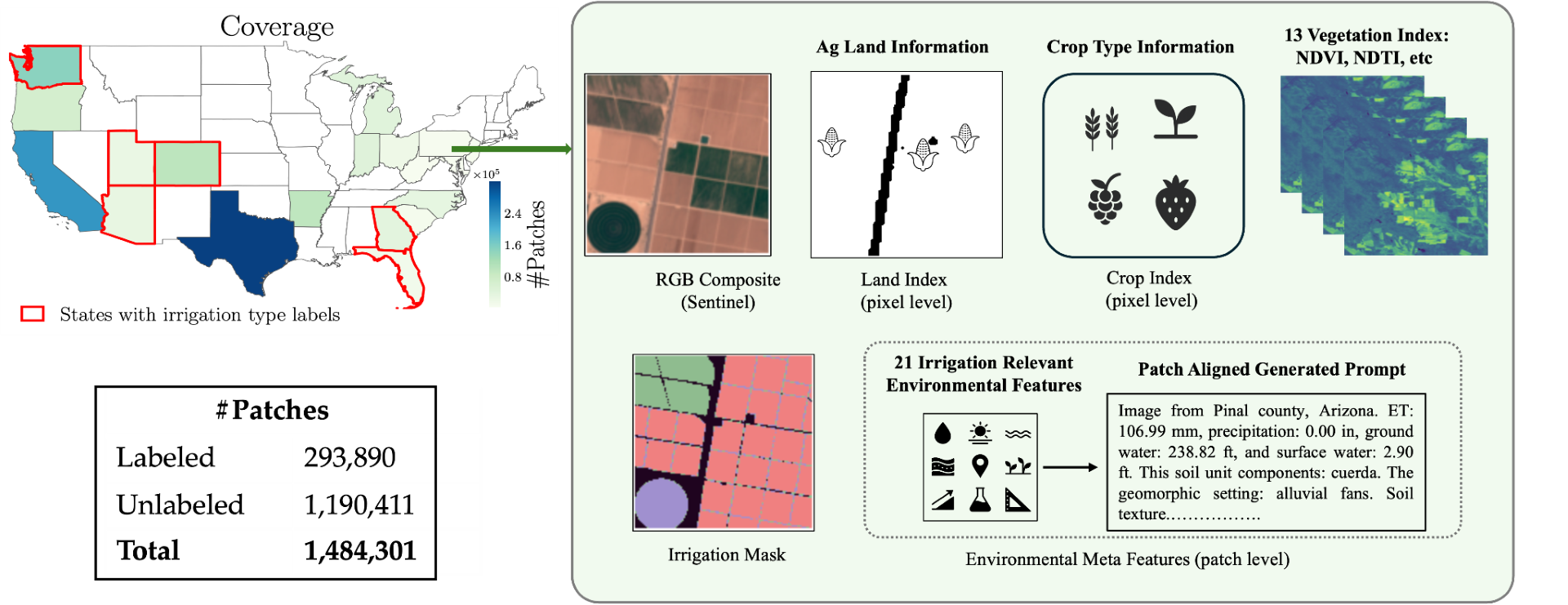 Figure 1. Graphical presentation of spatial data included in IrrMap, the first large-scale dataset to aid in the development of irrigation classifiers.
Figure 1. Graphical presentation of spatial data included in IrrMap, the first large-scale dataset to aid in the development of irrigation classifiers.
News
New GPUs available on Atlas
Twelve new graphics processing unit (GPU) nodes with a total of 48 GPUs are now available to all SCINet users on SCINet’s Atlas supercomputer. These nodes each have 4 NVIDIA L40S GPUs, 64 CPU cores, and 1.5 TB of memory. The L40S GPU features 48 GB of GPU memory and was designed for general-purpose workloads, including deep learning and AI applications. These new nodes are available in the “gpu-l40s” partition/queue on Atlas. Many thanks to the SCINet users who provided early testing and feedback for these new GPUs!
New Atlas storage and other upgrades
Atlas’s July 8 maintenance included replacing Atlas’s original data storage hardware with a completely new data storage system. This new system brings several important improvements, including:
- Nearly 3.4 times more storage space (increased from ~2.2 PB to ~9.6 PB).
- All solid-state storage that provides read/write performance superior to the previous storage system. This should be especially beneficial for data-intensive GPU workloads.
- Significant power and space savings compared to the previous storage system.
All files in /home and /project were automatically migrated to the new storage system. However, files in /90daydata were not automatically migrated. Instead, the old /90daydata is now available at /90daydata-old (read-only), and /90daydata is now located on the new storage system. If you have any files in /90daydata-old that you wish to retain, please copy them to a location on the new storage system. Otherwise, all files in /90daydata-old will expire as usual after 90 days of inactivity, after which the old storage system will be decommissioned.
Another change for Atlas made during the maintenance event was updating the GPU nodes’ time limits. To help promote resource sharing, jobs submitted to each of the GPU partitions (“gpu-v100”, “gpu-a100”, “gpu-a100-mig7”, and “gpu-l40s”) now have a maximum running time of 2 days.
Partition/queue changes on Ceres
During Ceres’ February 2025 maintenance, a new partition/queue, called “ceres”, was added to the supercomputer. This partition includes all community (i.e., non-priority) nodes and was intended to eventually replace all legacy community partitions. During Ceres’ June maintenance, the multitude of legacy community partitions (“short”, “medium”, “long”, “long60”, etc.) were removed, and nearly all compute jobs on Ceres should now be submitted to the “ceres” partition. Use of this partition has several benefits, including:
- Placing all Ceres community nodes into a single partition will result in shorter wait times and better cluster utilization.
- A simpler partition scheme makes Ceres easier and less confusing to use.
- The “ceres” partition is analogous to the “atlas” partition on Atlas and makes the user experience on both systems more similar.
Please note that the “ceres” partition has a default job time of 2 hours. This will help avoid long wait times in the queue due to implicitly requesting far more time than a job needs, but it also means that if your job requires more than 2 hours, you will need to explicitly request more time.
Also note that compute jobs in the “ceres” partition are limited to 3 weeks of run time by default. If you require very long run times, you may submit jobs using the “long” QOS (“quality of service”) specification. This will allow jobs to run for up to 60 days, but they will be limited to using no more than 144 CPU cores.
For more information, please see the SCINet website and the earlier announcement in our April newsletter.
New nodes on Ceres
The Ceres maintenance in June included a significant computing hardware upgrade for the supercomputer. All nodes on Ceres that were purchased in 2019 were decommissioned and replaced with 50 new nodes, each of which features 256 logical CPU cores and 2.3 TB of memory. These new nodes, along with the 20 new nodes added last year, not only have far more CPU cores than previous nodes, they also offer more memory than the earlier “high memory” nodes on Ceres. These nodes are available to all users in the “ceres” partition/queue and will, we hope, open up new computing opportunities for SCINet users.
Internships update
Our 2025 AI-COE/SCINet Graduate Student Internships Program summer interns are busy wrapping up their research projects with their ARS mentors!
We had 15 graduate students participate in summer or spring internships with ARS researchers this year. This year’s interns were affiliated with the New College of Florida, the University of Florida, and North Carolina State University. Participants were selected by their host institutions in part because of their expertise in computer science, machine learning (ML), and data science, and were paired with ARS researchers for projects that could benefit from the intern’s computational skills.
To recognize the accomplishments of these students, we held our annual internships research symposium on Monday, August 11, 2025. The symposium highlighted many inspiring applications of data science, AI, and ML methods to ARS research projects. We encourage you to watch the symposium recording to learn more!
Many thanks to the students who dedicated their time and hard work to these internships, the ARS scientists who volunteered to serve as internship mentors, and the universities who have partnered with us for this internships program.
Rangeland Analysis Platform datasets available via SCINet
The Rangeland Analysis Platform (RAP) provides remote- sensing- based vegetation cover and production estimates across the continental United States (1986-present). RAP data are helping to support producers through forage production and grazing management tools and to address some of the most pressing issues facing U.S. rangelands such as woody plant encroachment, invasive plants, drought, wind erosion, and fire. A host of federal agencies, organizations, scientists, and others are already using RAP to inform management strategies and monitor outcomes.
ARS scientists and collaborators can now access the RAP data on SCINet’s Ceres supercomputer through the Geospatial Common Data Library (GeoCDL) API. Currently, the annual 30-m vegetation cover (perennial forbs and grasses, annual forbs and grasses, shrubs, trees, litter, and bare ground) and annual herbaceous and perennial herbaceous production data as well as the 16-day annual and perennial herbaceous production data are available on Ceres. Soon we will also have a 10-m data product which includes the vegetation cover groups we have in the 30-m plus new invasive annual grass, sagebrush, pinyon-juniper, and canopy gap layers available through the GeoCDL API.
SCINet Working Groups
SCINet working groups (WGs) support ARS researchers and their collaborators in using scientific computing methods and SCINet computational resources in their research. Common WG activities include hosting recurring virtual meetings and webinars, organizing training events, and participating in collaborative research or software development projects.
Current Working Groups
- Ag100Pest Initiative (subgroup of AGR)
- Arthropod Genomics Research (AGR) Working Group
- Breeding AI and ML Working Group
- Geospatial Research Working Group
- Microbiome Working Group
- SCINet-Longterm Agroecosystem Research (LTAR) Phenology Working Group
- Protein Function and Phenotype Prediction Working Group
- Translational Omics Working Group
If you are interested in creating a working group, please compile the following:
- The working group’s name
- A description of the working group including its purpose and goals
- Contact information for people to reach out to if they want to learn more about or join the working group.
Send this information to the SCINet office at ARS-SCINet-Office@usda.gov.
Training
Training workshops
Practicum AI
Lead: Research Computing team at the University of Florida
Developed and presented by the University of Florida and customized for USDA-ARS with funding from ARS’s AI-COE, Practicum AI is a hands-on applied artificial intelligence curriculum intended for learners with limited coding and math background. The goal of this workshop series is to provide a streamlined introduction to the basic tools needed to train and use deep learning models. After completing these workshops, you will be ready to start experimenting with AI and to take more in-depth training, such as SCINet’s machine learning for science workshop series. The Carpentries workshop series offered in October (see below) will also provide a deeper exploration of several of the topics covered in this Practicum AI series.
Upcoming Practicum AI courses:
- Computing for AI: August 19, 2025, 2-5 PM ET
- Python for AI: August 27 & 28, 2025, 2-5 PM ET
- Deep Learning Foundations: September 3 & 4, 2025, 2 – 5 PM ET
Please note that the first two courses in this series cover tools and programming skills that are needed to use and train AI models. Course three (“Deep Learning Foundations”) will cover actual model training and implementation.
To register, please complete this registration form.
RNA-seq analysis with Galaxy
October 6 & 8, 2025, 1-5 PM ET
Leads: Genome Informatics Facility at Iowa State University and SCINet Office
In this workshop, participants will work through a complete RNA-seq analysis using SCINet’s Galaxy interface. Participants will learn how to create workflows in Galaxy to:
- Upload and process RNA-seq data.
- Perform quality control, alignment, and expression quantification.
- Identify differentially expressed genes using DESeq2.
We will also explore ways in which you can share your history and workflows with other SCINet users in Galaxy and how it can be a powerful tool for collaborations.
To register, please complete this registration form.
Carpentries
Leads: Keo Corak (ARS Computational Biologist), Amisha Poret-Peterson (ARS Research Microbiologist), and Steven Schroeder (ARS Computational Biologist)
The SCINet Office, in collaboration with ARS’s certified Carpentries instructors, is offering a Carpentries workshop that will teach participants the Unix command line, version control with Git, and Python programming. The workshop will span two weeks:
- Unix command line and version control with Git: October 27 & 29, 2025, 1 – 5 PM ET
- Programming with Python: November 3 & 5, 2025, 1 – 5 PM ET
This workshop will provide an interactive, hands-on experience that will help you learn valuable skills for data management and analysis. Please note that you may register for either week 1, week 2, or both, depending on which skills you’d like to learn.
To register for the Unix, Git, and Python Carpentries Workshop, please fill out this form.
Coursera
The SCINet Office and the AI-COE are excited to provide training opportunities through Coursera. Coursera licenses are available to ARS scientists and support staff for training focused on scientific computing, data science, artificial intelligence, and related topics. Successful completion of courses and specializations result in widely recognized certificates and credentials.
Please visit the SCINet Coursera Training Page to request a license. Licenses will be assigned on a rolling basis and are active for three months. Users may be able to extend their licenses upon request.
Workshop Reports
Foundations in bioinformatics workshop series
Leads: Genome Informatics Facility at Iowa State University and SCINet Office
Our Foundations in bioinformatics workshop series was held this past quarter and consisted of six workshops:
- Introduction to Bioinformatics: April 15, 2025
- Introduction to HPC Environments and Project Management and Organization: April 17, 2025
- Data Preparation and Quality Assessment in Genome Assembly: April 29, 2025
- Genome Assembly Validation and Improvement: May 1, 2025
- Introduction to RNA-seq Analysis: May 13, 2025
- RNA for Genome Annotation and Reproducibility in Bioinformatics: May 15, 2025
Throughout this series, participants gained hands-on experience in command-line tools for assessing the quality of sequencing data, genome assembly, annotation, and RNA-seq analysis. They also learned how to navigate high-performance computing environments and manage biological data effectively for reproducible research. Due to the high interest in this series, we will be offering these workshops again. Click here to join our waitlist!
From reads to variants: a pipeline for variant calling using DeepVariant
Leads: Sheina Sim (ARS Research Biologist), Craig Carlson (ARS Research Geneticist), and Haley Arnold (SCINet/AI-COE fellow)
This workshop provided a hands-on walkthrough of a variant-calling workflow using DeepVariant, a deep-learning based model for highly accurate variant calling. Participants learned how to process whole-genome sequencing data from multiple individuals by trimming and filtering raw reads, mapping to a reference assembly, and calling variants per individual. Participants also learned how to merge and filter variant call form (.vcf) files. The workshop included step-by-step guidance and best practices for variant calling on the SCINet high-performance computing clusters.
Click here to access this workshop’s materials and recording.
Automate your SCINet pipeline with Snakemake
Lead: Aaron Yerke (SCINet/AI-COE fellow)
This workshop introduced participants to Snakemake, a popular workflow management tool that supports documentation, organization, and reproducibility of computational workflows. Participants learned the basics of a Snakemake workflow and how to use it on SCINet clusters through a live demonstration of the pipeline. There was also an additional day of office hours for one-on-one integration of Snakemake into individual workflows/projects.
Click here to access this workshop’s materials.
Please help us improve our training offerings!
What scientific computing training do you need? The SCINet Office’s goal is to provide training opportunities and resources that meet the needs of ARS researchers, so we would be grateful if you could complete our short training request form and let us know how we can best help you learn the computing skills you need. Your feedback will help us decide where we should focus our efforts over the next year and beyond.
Training opportunities are continually being updated on the SCINet Upcoming Events webpage. For more information on any of the above trainings, registration questions, or suggestions, please email SCINet-training@usda.gov.
Support
Getting Started with SCINet is as easy as 1,2,3
If you do not already have a SCINet account, we hope you will consider joining the 2,300+ researchers who do. Follow the steps below to get started with SCINet.

- Request a SCINet account to gain access to computational and training resources.
- Read the SCINet FAQs covering helpful topics such as account management, accessing and installing software, obtaining storage space for your project(s), and how to get technical help.
- Visit the SCINet Forum to connect to other users, ask questions, and learn how SCINet can enable your research. P.S. Don’t forget to complete your annual USDA information security awareness training! This is required to maintain your account. For technical assistance with your SCINet account, please email scinet_vrsc@usda.gov.
Support email addresses
All requests for help with user accounts, login problems, resource requests, or support for the Ceres HPC cluster should be sent to the SCINet Virtual Research Support Core (VRSC) at scinet_vrsc@usda.gov. Help requests specific to the Atlas HPC cluster should be sent to help-usda@hpc.msstate.edu.
Many emails are currently being sent to other SCINet email inboxes. For the most expedient response to your support requests, be sure to send them to scinet_vrsc@usda.gov or to help-usda@hpc.msstate.edu for Atlas-specific requests.
SCINet User Tip
Requesting a specific node type
SCINet’s Ceres supercomputer has a variety of node types, each with different capabilities. For example, some nodes have AMD Epyc processors, others have Intel Xeon processors. In many cases, you can simply submit your compute jobs to the “ceres” partition/queue without worrying about these hardware details. However, what if you need to ensure that your job runs on a particular “kind” of node? For that, you can use Slurm’s “constraints” feature. Constraint flags allow you to tell Slurm exactly which type of node you need.
As an example, Ceres nodes with AMD Epyc 9754 processors have the “AMD” and “EPYC9754” constraints. To ensure your compute job runs on one (or more) of these nodes, use Slurm’s “–constraint=” option. You could, for instance, add the following to your sbatch script:
#SBATCH --constraint=EPYC9754
Again, most of the time, you don’t need to worry about this at all. But if you do need to leverage the capabilities of a particular kind of hardware, it is very handy! Please see the SCINet website for a table of all node types on Ceres and their associated constraint flags.
Do you have tips to share? Email them to ARS-SCINet-Office@usda.gov to be included in future newsletters.
SCINet Corner
SCINet Corner is a VRSC-moderated virtual space for people to share knowledge, discuss best practices, learn about new opportunities, and explore resources to support progress on their projects.
The next SCINet Corner will be held on August 28, 2025, from 1 – 2 PM ET. August’s event will introduce SCINet’s Galaxy interface, a web-based platform for data-intensive bioinformatics analyses.
You can register for this and future SCINet Corners here.
Have a question that just can’t wait? Want to see what other users are doing? Reach out to the ever-expanding SCINet Forum community for ideas, support, or just someone to bounce ideas off of at https://forum.scinet.usda.gov/.
Connect
The SCINet Community
To see all the SCINet community updates and review past newsletters, visit the Newsletter Archive.
Contribute
Do you use SCINet for your research? We would love to share your story! Email ARS-SCINet-Office@usda.gov to contribute content, ask questions, or provide feedback on the SCINet newsletter or website.
SCINet Office
Haitao Huang, Computational Biologist
Moe Richert, Web Developer
Lavida Rogers, Training Coordinator
Heather Savoy, Computational Biologist
Brian Stucky, Computational Biologist, Acting Chief Scientific Information Officer
SCINet Leadership Team
Brian Stucky, Acting Chief Scientific Information Officer
Rob Butler, SCINet Program Manager
Jeremy Edwards, Science Advisory Committee (SAC) Chair
Jeff Silverstein, Associate Administrator

